RAD-seq bioinformatics workshop
Malaga, Spain 2019
Part 2: Reproducible RAD-seq analyses
Deren Eaton, Columbia Univesity
Isaac Overcast, City College of New York
Objectives of this workshop
By the end of class you should be able to:
1. Branching: How to assemble multiple data sets under different parameter settings.
2. How to use the ipyrad API (Python).
4. Creating reproducible notebooks and sharing them.
Jupyter is a program that can be started from the command line.
It starts a server (the hub) that will open in your browser (you've seen this). From there, you can start notebooks that run a kernel (e.g., Python session) which you can interact with through the browser.
# Starting jupyter from the command line
$ jupyter-notebook
Starting a Python notebook in jupyter
Select [New] and then [Python 3]
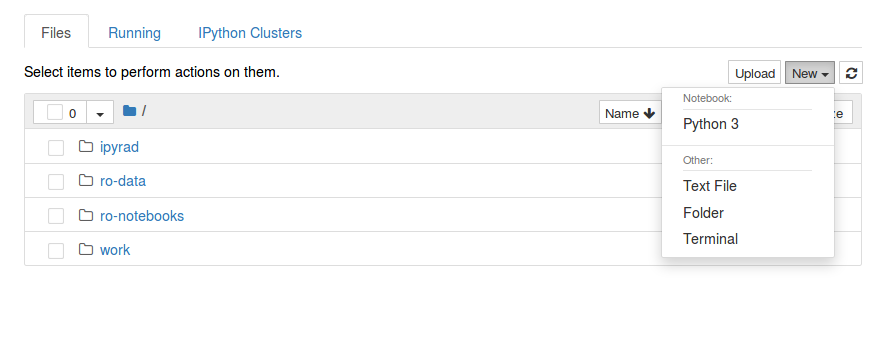
Getting started with notebooks
Code cells (Python), Markdown cells (rich text)
Getting started with notebooks
Code cells (Python), Markdown cells (rich text)
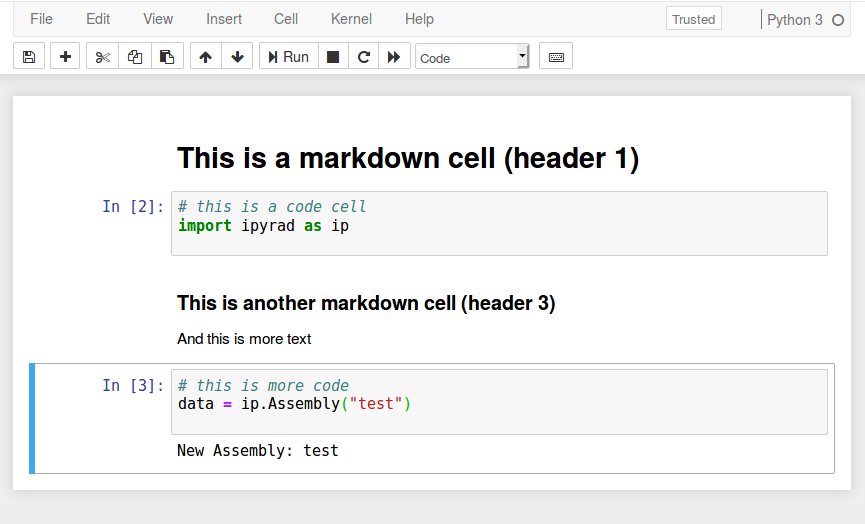
ipyrad CLI versus API
In the ipyrad CLI we use the '-n' flag
# CLI (terminal): create a new Assembly named "test"
$ ipyrad -n test
In the ipyrad API we create an Assembly object
# API (Python session): create a new Assembly named "test"
$ data = ip.Assembly("test")
Advantages to using the API
The API allows us to interactively set parameters, run assembly, etc.
# create the Assembly object
data = ip.Assembly("test")
# show the params
data.params
0 assembly_name test
1 project_dir ~
2 raw_fastq_path
3 barcodes_path
4 sorted_fastq_path
5 assembly_method denovo
6 reference_sequence
7 datatype rad
8 restriction_overhang ('TGCAG', '')
9 max_low_qual_bases 5
10 phred_Qscore_offset 33
...
Advantages to using the API
Setting params can take advantage of tab-completion
# set some parameters and try typing the first part and hitting [tab]
data.params.raw_fastq_path = "/home/jovyan/ro-data/ipsimdata/gbs_example_R1_.fastq.gz"
data.params.barcodes_path = "/home/jovyan/ro-data/ipsimdata/gbs_example_R1_.fastq.gz"
data.params.datatype = "gbs"
data.params
0 assembly_name test
1 project_dir ~
2 raw_fastq_path /home/jovyan/ro-data/ipsimdata/gbs_example_R1_.fastq.gz
3 barcodes_path /home/jovyan/ro-data/ipsimdata/gbs_barcodes.fastq.gz
4 sorted_fastq_path
5 assembly_method denovo
6 reference_sequence
7 datatype gbs
8 restriction_overhang ('TGCAG', '')
9 max_low_qual_bases 5
10 phred_Qscore_offset 33
...
Branching: Multiple assemblies done easily
The seven steps of assembly
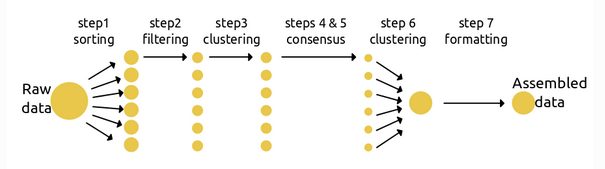
Branching: Multiple assemblies done easily
Start a different assembly from a previous checkpoint
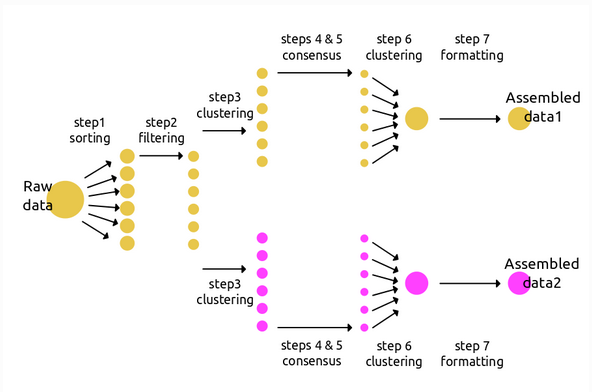
Branching: Multiple assemblies done easily
Create a new branch from an existing assembly.
# branching in the CLI
$ ipyrad -p params-simdata.txt -b newdata
loading Assembly: simdata
from saved path: ~/Documents/ipyrad/tests/simdata.json
creating a new branch called 'newdata' with 12 Samples
writing new params file to params-newdata.txt
Branching to drop samples
Create a new branch from an existing assembly.
# branching in the CLI
$ ipyrad -p params-simdata.txt -b newdata - 1A_0 1B_0
loading Assembly: simdata
from saved path: ~/Documents/ipyrad/tests/simdata.json
dropping 2 samples
creating a new branch called 'newdata' with 10 Samples
writing new params file to params-newdata.txt
Branching in the API
Create a new branch from an existing assembly.
# branching in the API
data1 = ip.Assembly("data1")
data1.params.clust_threshold = 0.85
# create a branch with different params
data2 = data1.branch("data2")
data2.params.clust_threshold = 0.90
# run both assemblies through steps 1-7
data1.run("1234567")
data2.run("1234567")
API for analysis
Even if data was assembled w/ CLI, API can be useful afterwards.
# load the assembly in the API
data = ip.load_json("/home/jovyan/work/simdata.json")
# show stats of the assembly
data.stats
# show output file paths of the assembly
data.outfiles
Phylogenomic analyses
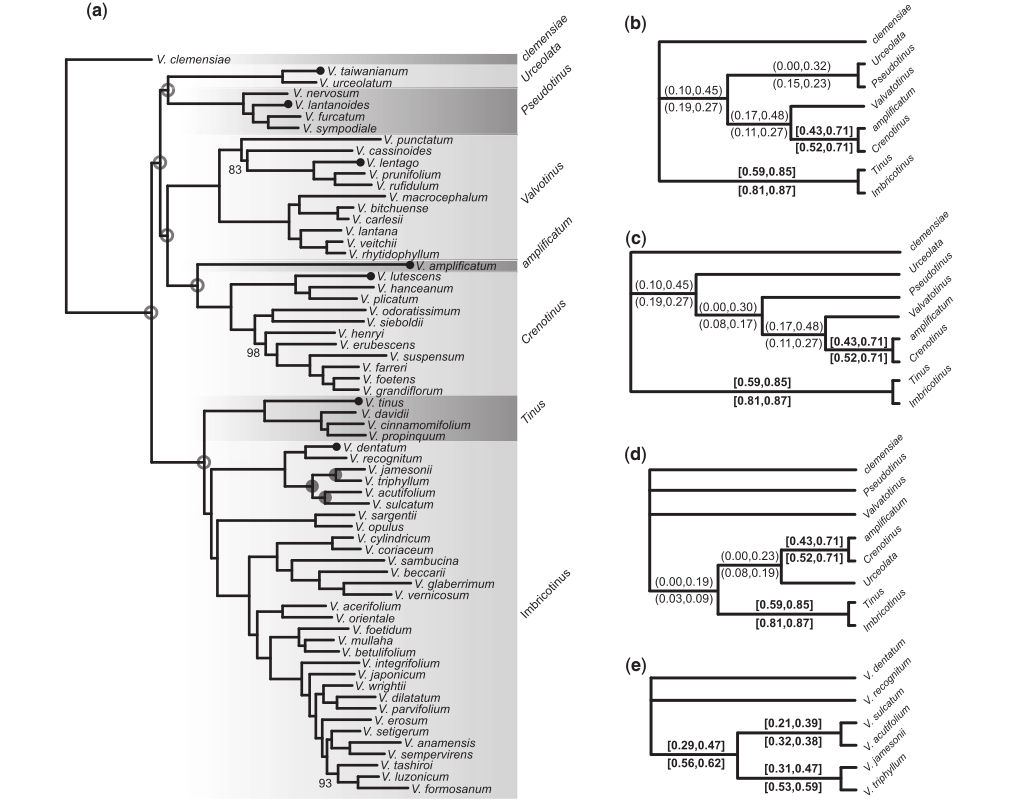
Infer gene trees and species trees, even over relatively deep evolutionary time scales (~100 Ma).
API analysis tools for population level analysis
Many other tools can be used with the output files as well.
# load the ipyrad analysis tools
import ipyrad.analysis as ipa
# run raxml with ipa
rax = ipa.raxml(
data="./simdata_outfiles/simdata.phy",
name="raxml-tree",
N=50,
T=4,
)
# run the analysis
rax.run()
Phylogenomic analyses
Infer gene trees and species trees from RAD-seq data.
Example github repositories with reproducible notebooks
https://github.com/dereneaton/Canarium-GBShttps://nbviewer.jupyter.org/github/dereneaton/ipyrad/blob/master/tests/cookbook-empirical-API-1-pedicularis.ipynb