Principles and Applications of Modern
DNA Sequencing
EEEB GU4055
Session 5: Homology & BLAST
Today's topics
1. Review notebook assignments: Numpy, Pandas.
2. BLAST and Homology.
3. OrthoDB and databases.
Homology and BLAST
What do we mean by orthology and paralogy?
Homology and BLAST
What do we mean by orthology and paralogy?Homology and BLAST
Question [1]: Using orthology to transfer the gene functional annotation from one organism to another might be inaccurate in some instances. What type of things do you think could lead to these inaccuracies? In other words, why is the function of the model organism gene not always the same as some other organism being compared to it?
OrthoDB
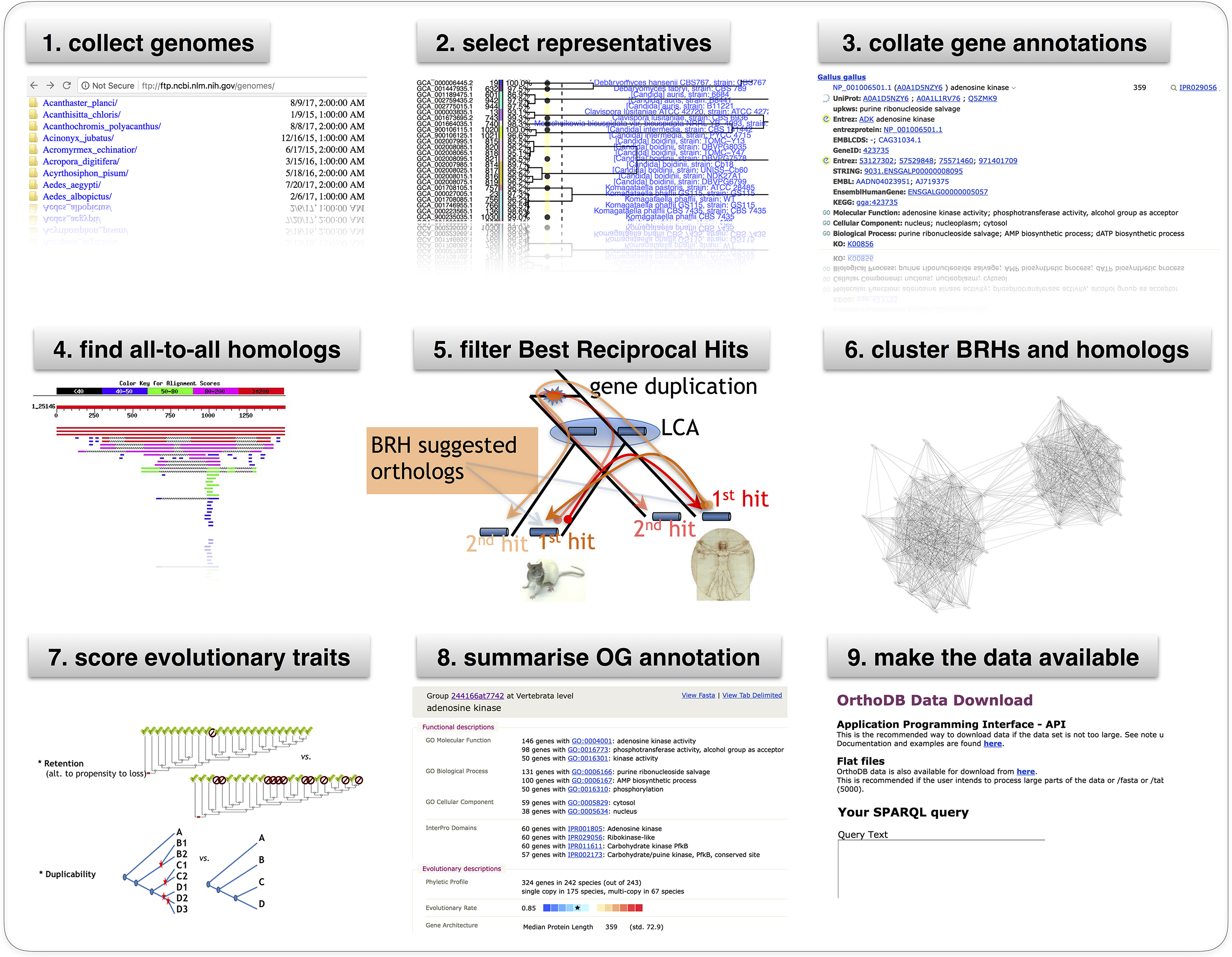
BLAST, NCBI, and APIs
BLAST is an algorithm for comparing sequences, it searches the NCBI database. The NCBI Entrez tools provide an API interface to this database. These tools have facilitated the development of complex tools like OrthoDB.

Using requests to query APIs
import requests
# search term
term = "FOXP2[GENE] AND Mammalia[ORGN] AND phylogenetic study[PROP]"
# make a request to esearch
res = requests.get(
url="https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi",
params={
"db": "nucleotide",
"term": term,
"sort": "Organism Name",
"retmode": "text",
"retmax": "20",
"tool": "genomics-course",
"email": "de2356@columbia.edu",
},
)